Research in the Marty lab focuses on developing cutting-edge mass spectrometry (MS) methods to study interactions with and within lipid membranes. We are especially interested in developing methods to study membrane protein-lipid interactions, mechanisms of membrane-active antimicrobial peptides, and new software approaches for native MS data analysis.
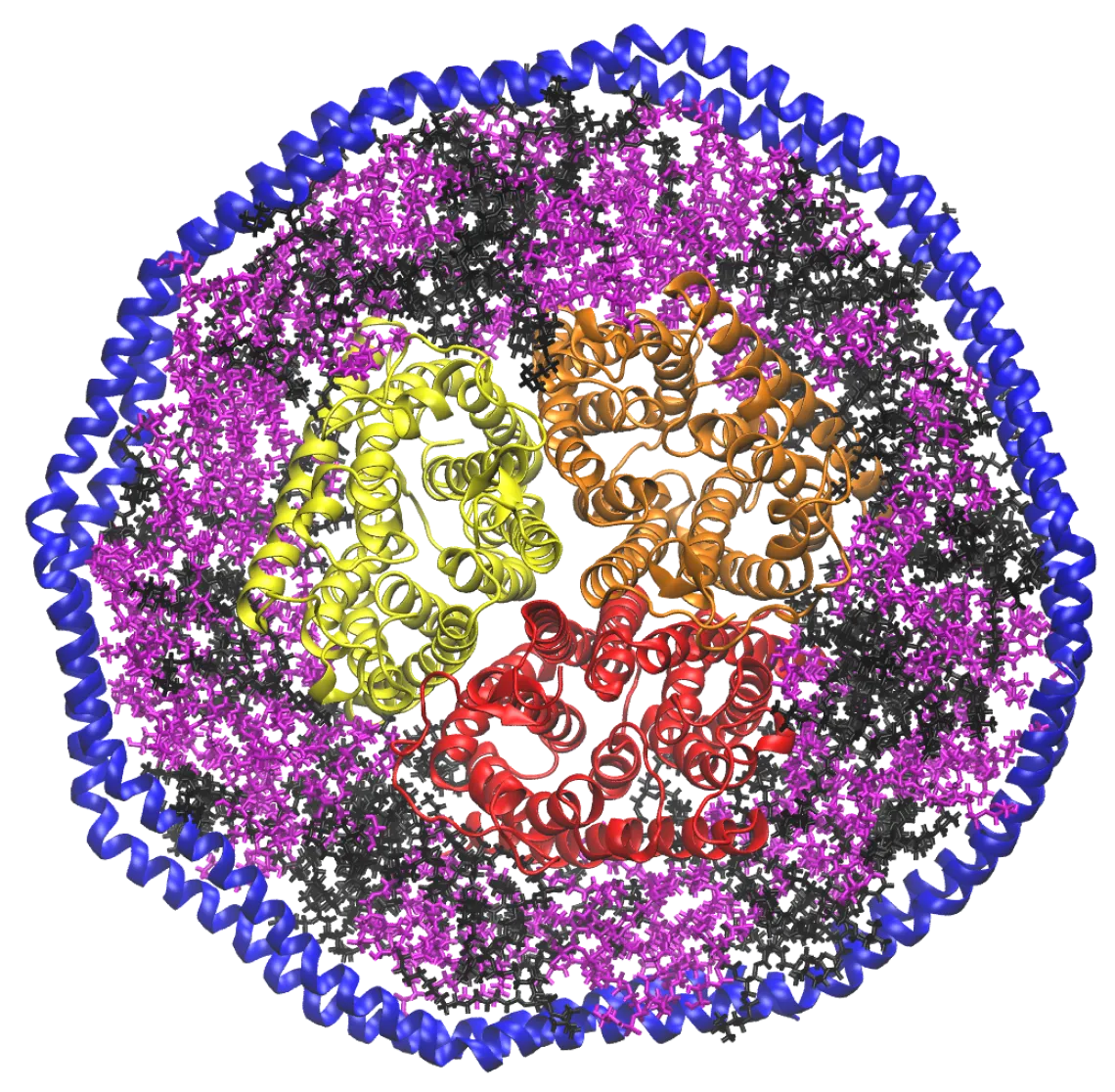
Membrane Protein-Lipid Interactions
Membrane proteins play a number of critical biochemical roles and make up the majority of drug targets. Despite their importance, membrane proteins remain challenging systems for analysis. The lipid bilayer can play an important but largely unexplored role in regulating membrane protein structure and function. New analytical and biochemical methods are necessary to better understand and design drugs to target membrane proteins.
Research in the Marty lab is focused on developing mass spectrometry methods to study the structure and biophysics of membrane proteins. Working at the interface of analytical chemistry and biochemistry, we use lipoprotein Nanodiscs to solubilize membrane proteins in a lipid bilayer with a defined composition. Nanodiscs are nanoscale discoidal lipid bilayers encircled by two amphipathic membrane scaffold proteins.
By using nondenaturing nano-electrospray ionization, we can keep the Nanodisc complex intact inside the mass spectrometer and gradually release the membrane protein with collisional activation. This approach, known as noncovalent or native mass spectrometry, allows us to measure the stoichiometry and lipid composition in large protein-lipid complexes to better understand protein-lipid interactions in membrane protein receptors and transporters.
We also use lipidomic analysis to identify and quantify lipids present in the nanodiscs. By examining exchange of lipids between nanodiscs with and without a target membrane protein, we can identify the local lipidome surrounding the protein.

Membrane Interacting Proteins and Peptides
Antimicrobial peptides (AMPs) are produced by all classes of life and present a promising approach to combat antibiotic resistance by targeting bacterial cell membranes. However, the fundamental chemical mechanisms of how AMPs interact with lipid membranes remains poorly understood. By assembling AMP complexes into Nanodiscs, we can use native MS to monitor the stoichiometry of peptides incorporated into different lipid bilayers and at increasing concentrations. A wide range of behaviors are observed, including strong differences in lipid and oligomeric specificity.
We are also applying these methods to study oligomerization of other protein and peptide targets. New research on viral ion channels (viroporins) reveals complex oligomerization behavior that depends on the external lipid and chemical environment. We have also used these techniques to study oligomerization and membrane interactions of amyloid-forming proteins involved in neurodegenerative diseases and diabetes. Our unique technology provides a powerful approach to study complex oligomerization of proteins and peptides when interacting with lipid bilayers.
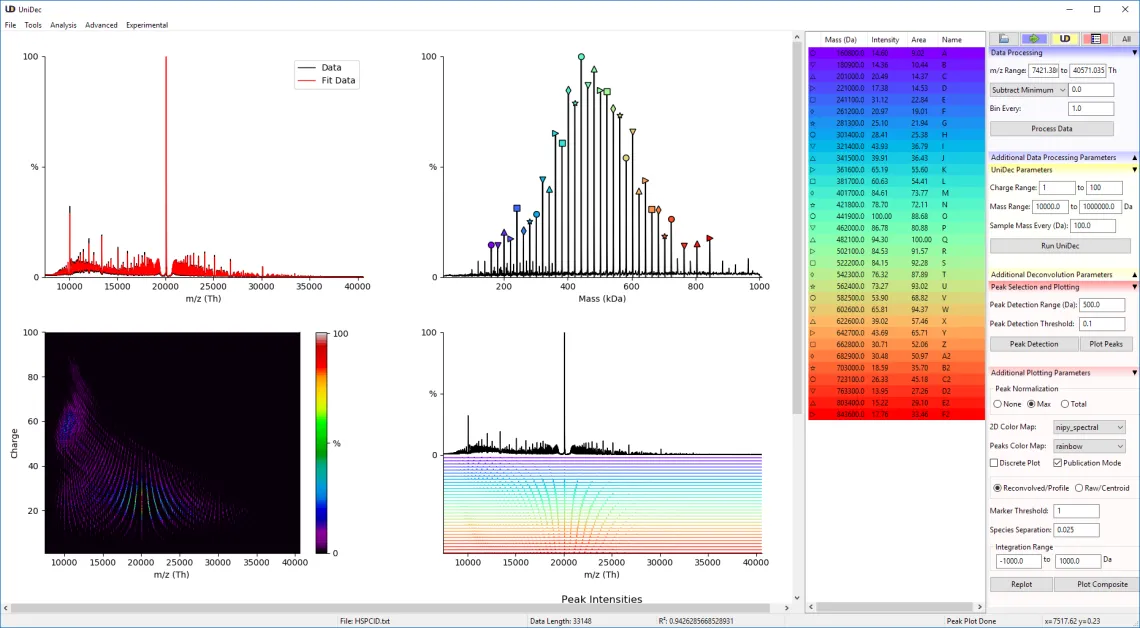
Native Mass Spectrometry Deconvolution
Because mass spectra of Nanodiscs are highly complex, we are developing computational approaches and software for analysis of native MS data. This work builds on UniDec software we have developed to rapidly and robustly deconvolve mass and ion mobility spectra along with CD-MS data. We have several avenues of active software development to improve the UniDec workflow and algorithms. UniDec has been downloaded over ten thousands times and used in several hundred published papers. It is also used by pharmaceutical companies for a range of applications.
